External users: registration to be carried out only through I-STEM portal
Additional information about sample and analysis details should be filled in the pdf form provided in the I-STEM portal under “DOWNLOAD CSRF”
Internal users (IITB): registration to be carried out only through DRONA portal
Additional information about sample and analysis details should be filled in the pdf form provided here.
.
Category
- Microscopy and Imaging » Force Microscopy
- Material Characterization » Electrical Characterisation
Booking Details
Facility Management Team and Location
Facility Features, Working Principle and Specifications
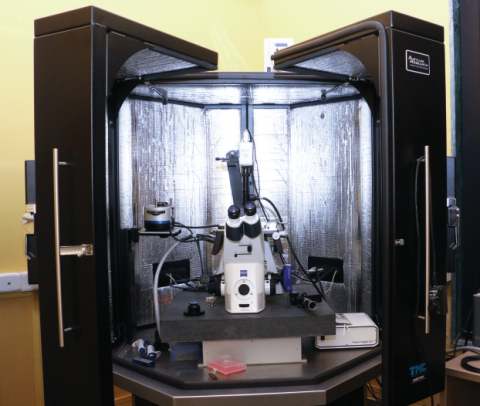
Facility Description
The Bio-AFM facility was installed in January 2014, in the department of “Bio-Science & Bio-Engineering” Central Facility as per RIFC norms. The Facility is open for all IIT Bombay internal users, other institute, National Laboratory and Industry.
The principle of operation of the AFM is very simple - A sharp cantilever tip interacts with the sample surface sensing the local forces between the molecules of the tip and sample surface. This instrument is not a “conventional microscope” that collects and focuses light. The word microscope has been associated with this instrument because it is able to measure microscopic features of the sample. The most characteristic property of the AFM is that the images are acquired by “feeling” the sample surface without using light. In this way, not only the sample topography can be recorded with good resolution, but also the material characteristics and the strength of interaction between the sample surface and the cantilever tip. Due to the fact that no light is involved in acquiring the sample properties, the AFM reaches a resolution far below the diffraction limit offered by the optical microscopy. Its resolution is limited only by the tip radius and the spring constant of the cantilever.
SYSTEM SPECIFICATION:
Closed loop sensors on all three axes:
X & Y range 120 μm, X & Y sensors <0.5 nm noise, <0.5% non-linearity
Z range > 40 μm
DC height noise <50 pm.
Lowest Noise Single Molecule or Cellular Force Measurements:
Cantilever deflection noise <15 pm (typical 8 pm)
Low coherence source Super luminescent diode (SLD) for ripple-free baseline.
Cantilever spring constant calibration by the thermal noise and Sader methods or GetReal automated cantilever calibration.
Flexible interface allows recording or triggering from any channel during a force curve, including amplitude/phase from AC or Dual AC™ mode; user-supplied input voltages; and photon count rate (with optional Digital Access Module).
Force mapping including automated adhesion and elastic modulus analysis.
SPECIAL FEATURES:
High-Resolution imaging in liquid for soft biological samples.
No pre-processing of materials/cells is required for imaging.
Ability to combine AFM measurements with images obtained in inverted microscope.
Real-time Optical Navigation Top or bottom-view optical images can be used to navigate the tip to any feature on the sample and then scan that area at the nanoscale with the AFM or select specific locations for force curves – easily and seamlessly.
Powerful Real-time and Offline Rendering Options Both AFM and optical images can be rendered and viewed together in both real-time and offline. Optical images can be overlaid on AFM data to assist interpretation. Stunning 3D renderings combine AFM topography with the capabilities of light microscopy.
All of the following optical techniques are supported:
1. Bright field
2. Phase Contrast
3. Fluorescence
Large Z range (40 μm extended Z option) accommodates demanding applications such as cell-cell and cell-substrate adhesion measurements.
Users can choose between open loop force curves with sensored Z for the ultimate in low noise performance or closed loop Z for the most accurate velocity control.
Force Mapping measures force-distance curves at a grid of points with automated fitting of indentation models for estimation of elastic modulus and automated adhesion /rupture force analysis.
Analysis software helps by suggesting the most appropriate indentation model among many built-in options, including “Hertz / Sneddon, Johnson-Kendall-Roberts (JKR), DerjaguinMüller-Toporov (DMT), and Oliver-Pharr”.
Sample Preparation, User Instructions and Precautionary Measures
- All type of samples can be done.
- Expect Suspended and semi liquid sample are not allowed
We will accept online registration only through the IRCC webpage. If you need to cancel your slot, please email us immediately with an explanation.
Slots will be provided on a first-come-first-served basis.
USB drives are strictly prohibited for copying data to minimize virus-related issues. You are requested to bring a new blank CD to transfer your data. All data must be transferred within 7 days of imaging. Without exception.
Users must be present during the entire slot.
Charges for Analytical Services in Different Categories
Basic Charges + GST*(as applicable).
GST* rate as on 1.8.2017:
- If the recipient of the report is from Maharashtra: 9% SGST & 9% CGST.
If the recipient of the report is from outside Maharashtra: 18% IGST.
Sr. No. | Category | Revised Charges |
1 | IITB (TA) | ₹ 750.00 |
2 | IITB STUDENT | ₹ 1,500.00 |
3 | IITB-Monash Students | ₹ 1,500.00 |
4 | Academic Institutes | ₹ 7,500.00 |
5 | National Labs | ₹ 7,500.00 |
6 | Sine (Letter from SINE reqd.) | ₹ 7,500.00 |
7 | Research Park (MSME) | ₹ 7,500.00 |
8 | Research Park (Big Industry partners) | ₹ 11,250.00 |
9 | Industries | ₹ 15,000.00 |
- You can make the payment ONLINE in favor of BIO-AFM Facility, IIT Bombay.
Bank details as follow,
Bank Name: State Bank of India, IIT Powai Branch, Mumbai-400076, India.
Bank A/c No.: 10725729173. IFSC/RTGS/NEFT: SBIN0001109.
MICR no. : 400002034. Branch Code: 1109.
Swift Code : SBININBB519. Telephone No. : +91-22-2159-6746.
Applications
Biological sciences.
Life science
Physical sciences
Material science
Polymer science
Electrical characterization
Nanolithography
Nanotechnology
Nano-mechanics.
Sample Details
Cell Media is allowed
All type of substrate are allowed
No need of gases
- Maximum 2” X 2” in x-y and Height 14 mm
- NA
- NA
- NA
SOP, Lab Policies and Other Details
Publications
2013 :
Ghosh, D., Mondal, M., Mohite, G. M., Singh, P. K., Ranjan, P., Anoop, A., Ghosh, S., Jha, N.N., Kumar, A. & Maji, S. K. (2013). "The Parkinson's disease-associated H50Q mutation accelerates α-Synuclein aggregation in vitro." Biochemistry, 52(40), 6925- 6927
2014 :
D Ghosh, S Sahay, P Ranjan, S Salot, GM Mohite, PK Singh, S Dwivedi, E Carvalho, R Banerjee, A Kumar and S.K. Maji (2014) "The newly Discovered Parkinson's Disease Associated Finnish Mutation (A53E) Attenuates α-Synuclein Aggregation and Membrane Binding," Biochemistry, 53(41):6419-21
2015 :
D Ghosh, PK Singh, S Sahay, NN Jha, RS Jacob, S Sen, A Kumar, R Riek and SK Maji (2015) "Structure based aggregation studies reveal the presence of helix-rich intermediate during α-Synuclein aggregation", Scientific Rep, 5:9228.
Reeba S. J.*, George E*, Singh P. K., Salot S., Anoop A., Jha N. N., Sen S.# and Maji S. K.#, "Cell adhesion on amyloid fibrils lacking integrin recognition motif", J Biol Chem. 2016 Mar 4;291(10):5278-98.
Ranjan P, Kumar A. "The Involvement of His50 during Protein DisulfideIsomerase Binding Is Essential for Inhibiting α-Synd Fibril Formation.Biochemistry". 2016 May17; 55(19):2677-80
2017 :
L. K. Sthanam, A. Barai A. Rastogi, V. K. Mistari, A. Maria, R. Kauthale, M. Gatne, S. Sen, "Biophysical regulation of mouse embryonic stem cell fate and genomic integrity by feeder derived matrices", Biomaterials, Volume 119, March 2017, Pages 9-22
http://www.sciencedirect.com/science/article/pii/S0142961216306950?via
J. Rane, P. Bhaumik, D. Panda, "Curcumin inhibits tau aggregation and disintegrates tau filaments in vitro" Journal of Alzheimer's disease, 2017.
A. Yadav and M.S. Tirumkudulu, "Free-standing monolayer films of ordered colloidal particles", Soft Matter, 13, 4520-4525 (2017).
http://pubs.rsc.org/en/content/articlelanding/2017/sm/c7sm00407a#!divA
Ranjan, P., Ghosh, D., Yarramala, D. S., Das, S., Maji, S. K., & Kumar, A. (2017). "Differential copper binding to alpha-synuclein and its disease-associated mutants affect the aggregation and amyloid formation." Biochimica et Biophysica Acta (BBA)- General Subjects, 1861(2), 365-374.
http://www.sciencedirect.com/science/article/pii/S0304416516304822
Srinivasan S.*, Ashok V.*, Mohanty S., Das A., Das S., Kumar S., Sen S.#, Purwar R.#, "Blockade of Rho-associated protein kinase (ROCK) inhibits the contractility and invasion potential of cancer stem like cells", Oncotarget, 2017.
Dey S. K., Singh R. K., Chattoraj S., Saha S., Das A., Bhattacharyya K., Sengupta K., Sen S., Jana S. S., "Differential role of nonmuscle myosin II isoforms during blebbing of MCF-7 cells.", Mol. Biol. Cell., 2017.
2018:
Surendra Kumar Verma, Akshay Modi, Ashwain Dravid, Jayesh Bellare (2018). “Lactobionic acid-functionalized polyethersulfone hollow fiber membranes promote HepG2 attachment and functions.” RSC Advances, 8 (51) 29078-29088
Dubey R, Minj P, Malik N, Sardesai DM, Kulkarni SH, Acharya JD, Bhavesh NS, Sharma S, Kumar A (2017) “Recombinant human islet amyloid polypeptide forms shorter fibrils and mediates β-cell apoptosis via generation of oxidative stress.” Biochem J. 16:3915-3934
R Kumar, S Das, GM Mohite, SK Rout, S Halder, NN Jha, S Ray, S Mehra, V Agarwal and SK Maji (2018), “Cytotoxic oligomers and fibrils trapped in a gel-like state of α-synuclein assemblies.” Angewandte Chemie International Edition
Bhattacharya D, Sinha K, Panda D. “Mutation of G51 in SepF impairs FtsZ assembly promoting ability of SepF and retards the division of Mycobacterium smegmatis cells.” Biochem J. 2018 Aug 14;475(15):2473-2489.
S. Mehra, D Ghosh, R Kumar, M Mondal, LG Gadhe, S Das, A Anoop, NN Jha, RS Jacob, D Chatterjee, S Ray, N Singh, A Kumar, and SK Maji (2018), “Glycosaminoglycans have variable effects on α-synuclein aggregation and differentially affect the activities of the resulting amyloid fibrils.” Journal of Biological Chemistry.
GM Mohite, A Navalkar, R Kumar, S Mehra, S Das, LG Gadhe, D Ghosh, B Alias, V Chandrawanshi, A Ramakrishnan, S Mehra and SK Maji (2018), “Familial α-synuclein A53E mutation enhances cell death in response to environmental toxins due to more population of oligomers.” Biochemistry
S Das, MK Kumawat, S Ranganathan, R Kumar, J Adamcik, P Kadu, R Paadinhaateri, R Srivastava, R Mezzenga and SK Maji (2018), “Cell alignment on graphene-amyloid composites.” Advanced Materials Interfaces.
VENUGOPAL B, MOGHA P, DHAWAN J, & MAJUMDER A. (2018). “Cell density overrides the effect of substrate stiffness on human mesenchymal stem cells' morphology and proliferation.” Biomaterials Science. 6, 1109-1119.
Sthanam L.K., Saxena N., Mistari V., Roy T., Jadhav S., Sen S., “Initial priming on soft substrates enhances subsequent topography-induced neuronal differentiation in ESCs but not in MSCs,” ACS Biomaterials Science & Engineering, 2018.
Kumar, S., Das, A., Sen, S., “Multi-compartment cell-based modeling of confined migration: regulation by cell intrinsic and extrinsic factors,” Mol. Biol. Cell, 2018. 15.
George E., Barai A., Shirke P., Majumder A., Sen S., "Engineering Interfacial Migration by Collective Tuning of Adhesion Anisotropy and Stiffness", Acta Biomaterialia, 2018.
Saxena N., Mogha P., Dash S., Majumder A., Jadhav S.*, Sen S*., "Matrix elasticity regulates mesenchymal stem cell chemotaxis", J. Cell Sci. 2018
Kumar S*., Das A.*, Barai A., Sen S., "MMP secretion rate and inter-invadopodia spacing collectively govern cancer invasiveness", Biophysical Journal, February 2018.
Kapoor A.*, Barai A.*, Thakur B., Das A., Patwardhan S. R., Monteiro M., Gaikwad S., Bukhari A., Mogha P., Majumder A., De A., Ray P.#, Sen S.#, "Soft drug-resistant ovarian cancer cells migrate via two distinct mechanisms utilizing myosin II-based contractility", BBA Molecular Cell Research, February 2018.
Mundhara, N., Majumder, A., and Panda, D. (2019). “Methyl-β-cyclodextrin, an actin depolymerizer augments the antiproliferative potential of microtubule-targeting agents.” Scientific reports, 9(1), 7638.
Dhaked, H. P., Ray, S., Battaje, R. R., Banerjee, A. and Panda, D. (2019). Regulation of Streptococcus pneumoniae FtsZ assembly by divalent cations: paradoxical effects of Ca2+ on the nucleation and bundling of FtsZ polymers. FEBS J. 2019,
Rane, J. S., Kumari, A., and Panda, D. (2019). “An acetylation mimicking mutation, K274Q, in tau imparts neurotoxicity by enhancing tau aggregation and inhibiting tubulin polymerization.” Biochemical Journal, 476(10), 1401-1417.
Bhattacharya, D., Sinha, K., and Panda, D. (2018). “Mutation of G51 in SepF impairs FtsZ assembly promoting ability of SepF and retards the division of Mycobacterium smegmatis cells.” Biochemical Journal, 475(15). 2473-2489.
2019-2020:
Surendra Kumar Verma, Akshay Modi, Ashwain Dravid, Jayesh Bellare “Lactobionic acid-functionalized polyethersulfone hollow fiber membranes promote HepG2 attachment and functions”. RSC Advances, 8 (51) 29078-29088
Kumar, S., Das, A., Sen, S., “Multi-compartment cell-based modeling of confined migration: regulation by cell intrinsic and extrinsic factors,” Mol. Biol. Cell, 2018. 15.
George E., Barai A., Shirke P., Majumder A., Sen S., “Engineering Interfacial Migration by Collective Tuning of Adhesion Anisotropy and Stiffness", Acta Biomaterialia, 2019.
Mundhara, N., Majumder, A., and Panda, D. “Methyl-β-cyclodextrin, an actin depolymerizer augments the antiproliferative potential of microtubule-targeting agents.” Scientific reports, 9(1), 7638.
Dhaked, H. P., Ray, S., Battaje, R. R., Banerjee, A. and Panda, D. “Regulation of Streptococcus pneumoniaeFtsZ assembly by divalent cations: paradoxical effects of Ca2+ on the nucleation and bundling of FtsZ polymers.” FEBS J.,
Rane, J. S., Kumari, A., and Panda, D. “An acetylation mimicking mutation, K274Q, in tau imparts neurotoxicity by enhancing tau aggregation and inhibiting tubulin polymerization.” Biochemical Journal, 476(10), 1401-1417.
2020-21:
Rane, J.S., A. Kumari, and D. Panda, “The Acetyl Mimicking Mutation, K274Q in Tau, Enhances the Metal Binding Affinity of Tau and Reduces the Ability of Tau to Protect DNA.” ACS Chemical Neuroscience, 2020. 11(3): p. 291-303.
Rane, J.S., A. Kumari, and D. Panda,“An acetylation mimicking mutation, K274Q, in tau imparts neurotoxicity by enhancing tau aggregation and inhibiting tubulin polymerization.” Biochem J, 2019. 476(10): p. 1401-1417.
Chavan, S.S. and H.K. Bagla, “Alpha track detection study on CR-39 from granitic wastes employing tetraethyl ammonium bromide as chemical etchant.” Journal of Radioanalytical and Nuclear Chemistry, 2020. 325(3): p. 823-830.
https://link.springer.com/article/10.1007/s10967-020-07259-6
Siddiquie, R.Y., et al., “Anti-Biofouling Properties of Femtosecond Laser-Induced Submicron Topographies on Elastomeric Surfaces.” Langmuir, 2020. 36(19): p. 5349- 5358.
Poojari, R., et al., “Antihepatoma activity of multifunctional polymeric nanoparticles via inhibition of microtubules and tyrosine kinases.” Nanomedicine (Lond), 2020. 15(4): p. 381-396.
Dinda, S.K., S. Polepalli, and C.P. Rao, “Binding of Fe(ii)-complex of phenanthroline appended glycoconjugate with DNA, plasmid and an agglutinin protein.” New Journal of Chemistry, 2020. 44(27): p. 11727-11738.
https://pubs.rsc.org/en/content/articlelanding/2020/nj/d0nj01524e#!divRelatedContent&articles
Adhyapak, P., et al., “Dynamical Organization of Compositionally Distinct Inner and Outer Membrane Lipids of Mycobacteria.” Biophys J, 2020. 118(6): p. 1279-1291.
Sharma, K., et al., “Effect of Disease-Associated P123H and V70M Mutations on beta- Synuclein Fibrillation.” ACS Chem Neurosci, 2020. 11(18): p. 2836-2848.
Bhatia, E. and R. Banerjee, “Hybrid silver-gold nanoparticles suppress drug resistant polymicrobial biofilm formation and intracellular infection.” J Mater Chem B, 2020. 8(22): p. 4890-4898.
Shashank, B.S., et al., “Investigations on biosorption and biogenic calcite precipitation in sands.” Soil Use and Management, 2020. n/a(n/a).
Mundhara, N., A. Majumder, and D. Panda, “Methyl-beta-cyclodextrin, an actin depolymerizer augments the antiproliferative potential of microtubule-targeting agents.” Sci Rep, 2019. 9(1): p. 7638.
Mishra, M. and S. Kapoor, “Modulation of host cell membrane nano-environment by mycobacterial glycolipids: Involvement of PI(4,5)P2 signaling lipid?” Faraday Discussions, 2020.
Mishra, M., et al., “Mycobacterium Lipids Modulate Host Cell Membrane Mechanics, Lipid Diffusivity, and Cytoskeleton in a Virulence-Selective Manner.” ACS Infect Dis, 2020. 6(9): p. 2386-2399.
Mishra, M., et al., “Mycobacterium Lipids Modulate Host Cell Membrane Mechanics, Lipid Diffusivity, and Cytoskeleton in a Virulence-Selective Manner.” ACS Infect Dis, 2020. 6(9): p. 2386-2399.
Sadgar, A.L., T.S. Deore, and R.V. Jayaram, “Pickering Interfacial Catalysis- Knoevenagel Condensation in Magnesium Oxide-Stabilized Pickering Emulsion.” Acs Omega, 2020. 5(21): p. 12224-12235.
Behera, T., et al., “Spatially correlated photoluminescence blinking and flickering of hybrid-halide perovskite micro-rods.” Journal of Luminescence, 2020. 223: p. 117202.
Pratihar, S., et al., “Tailored piezoelectric performance of self-polarized PVDF-ZnO composites by optimization of aspect ratio of ZnO nanorods.” Polymer Composites, 2020. 41(8): p. 3351-3363.
Dadhich, R., et al., “A Virulence-Associated Glycolipid with Distinct Conformational Attributes: Impact on Lateral Organization of Host Plasma Membrane,” Autophagy, and Signaling. ACS Chem Biol, 2020. 15(3): p. 740-750.
Mehra, S., et al., “α-Synuclein aggregation intermediates form fibril polymorphs with distinct prion-like properties.” bioRxiv, 2020: p. 2020.05.03.074765.
Sushma S.C. , Hemlata K. B., “Comparative on alpha Track detection from phosphate fertilizer industrial effluent employing polymeric solid state Nuclear Track Detector.” Alochana Chakra Journal, Volume IX, Issue V, May/2020. ISSN No. 2231-3990.
Dutta, S., et al., “Chemical evidence of preserved collagen in 54-million-year-old fish vertebrae.” Palaeontology, 2020. 63(2): p. 195-202.
Kumari, P., A. Modi, and J. Bellare, “Enhanced flux and antifouling property on municipal wastewater of polyethersulfone hollow fiber membranes by embedding carboxylated multi-walled carbon nanotubes and a vitamin E derivative.” Separation and Purification Technology, 2020. 235: p. 116199.
Mukherjee, A., et al., “Nuclear Plasticity Increases Susceptibility to Damage During Confined Migration.” bioRxiv, 2020: p. 2020.01.18.911529.
2021-22:
Singh, D., P. Singh, A. Pradhan, R. Srivastava, and S.K. Sahoo, “Reprogramming Cancer Stem-like Cells with Nanoforskolin Enhances the Efficacy of Paclitaxel in Targeting Breast Cancer.” ACS Appl Bio Mater, 2021. 4(4): p. 3670-3685
Mukherjee, S. and D. Panda, “Contrasting Effects of Ferric and Ferrous Ions on Oligomerization and Droplet Formation of Tau: Implications in Tauopathies and Neurodegeneration.” ACS Chem Neurosci, 2021. 12(23): p. 4393-4405.
Pradhan, A., S. Mishra, A. Surolia, and D. Panda, “C1 Inhibits Liquid-Liquid Phase Separation and Oligomerization of Tau and Protects Neuroblastoma Cells against Toxic Tau Oligomers.” ACS Chem Neurosci, 2021. 12(11): p. 1989-2002.
Sthanam, LK, Roy T, Patwardhan S, Shukla A, Sharma S, Shinde PV, Kale HT, Shekar PC, Kondabagil K, Sen S*, “MMP modulated differentiation of mouse embryonic stem cells on engineered cell derived matrices”, Biomaterials, 2021, 121268.
Patwardhan S*, Mahadik P, Shetty O, Sen S*, “ECM stiffness-tuned exosomes drive breast cancer motility through thrombospondin-1”, Biomaterials, 2021, 279: 121185.
Barai A, Mukherjee A, Das A, Saxena N, and Sen S*, “α-ctinin-4 drives invasiveness by regulating myosin IIB expression and myosin IIA localization”, J. Cell Sci, 2021: jcs.25858.
Asadullah…., Kumar S # *, Saxena N, Sarkar M, Barai A, Sen S*, “Combined heterogeneity in cell size and deformability promotes cancer invasiveness”, J. Cell Sci., 2021, jcs.250225
Jahan I, Pandya J, Munshi R, Sen S*, “Glycocalyx disruption enhances motility, proliferation and collagen synthesis in diabetic fibroblasts”, BBA Mol. Cell Res., 2021, 1868(4):118955.
Shirke et al., “Viscotaxis”- Directed Migration of Mesenchymal Stem Cells in Response to Loss Modulus Gradient”, 2021, Acta Biomaterialia, 135, pp 356-367.
Mundhara, N., et al., “Hyperthermia induced disruption of mechanical balance leads to G1 arrest and senescence in cells”, 2021 Biochemical Journal, 478, pp 179–196.
Shetty, S., A.M. Shanmugharaj, and S. Anandhan, “Physico-chemical and piezoelectric characterization of electroactive nanofabrics based on functionalized graphene/talc nanolayers/PVDF for energy harvesting.” Journal of Polymer Research, 2021. 28(11): p. 419.
2022-23:
Venkatramani, A., S. Mukherjee, A. Kumari, and D. Panda, “Shikonin impedes phase separation and aggregation of tau and protects SH-SY5Y cells from the toxic effects of tau oligomers.” Int J Biol Macromol, 2022. 204: p. 19-33.
Mishra, M. and S. Kapoor, Chapter 5 – “Multifaceted roles of mycobacterium cell envelope glycolipids during host cell membrane interactions, in Biology of Mycobacterial Lipids,” Z. Fatima and S. Canaan, Editors. 2022, Academic Press. p. 105- 131.
Chowdhury, M., M. Madhusudanan, and J. Sarkar. “Conflicting role of plasticization in nanorheology of out-of-equilibrium thin polystyrene films.”
https://ui.adsabs.harvard.edu/abs/2022APS..MARA17005C/abstract
Adhyapak, P., et al., “Lipid Clustering in Mycobacterial Cell Envelope Layers Governs Spatially Resolved Solvation Dynamics.” Chemistry – An Asian Journal, 2022. 17(11): p. e202200146.
Singh, B., et al., “Fabrication and cytotoxicity evaluation of polyethyleneimine conjugated fluorescent MXene nanosheets as cancer theranostics agent.” Polymer Bulletin, 2022.
Menon, A.P., et al., “Mutually Exclusive Interactions of Rifabutin with Spatially Distinct Mycobacterial Cell Envelope Membrane Layers Offer Insights into Membrane-Centric Therapy of Infectious Diseases.” ACS Bio & Med Chem Au, 2022. 2(4): p. 395-408.
2023-24 :
Madhusudanan, M., et al., “Tuning the Plasticization to Decouple the Effect of Molecular Recoiling Stress from Modulus and Viscosity in Dewetting Thin Polystyrene Films.” Macromolecules, 2023. 56(4): p. 1402-1409.
Mogha, P., S. Iyer, and A. Majumder, “Extracellular matrix protein gelatin provides higher expansion, reduces size heterogeneity, and maintains cell stiffness in a long- term culture of mesenchymal stem cells.” (1532-3072 (Electronic)).
Singh, B., et al., “Preclinical safety assessment of red emissive gold nanocluster conjugated crumpled MXene nanosheets: a dynamic duo for image-guided photothermal therapy.” Nanoscale, 2023. 15(6): p. 2932-2947.
Singh, B., et al., “Synthesis and degradation mechanism of renally excretable gold core– shell nanoparticles for combined photothermal and photodynamic therapy.” Nanoscale, 2023. 15(3): p. 1273-1288.
Maity, S., A. Sasmal, and S. Sen, “Barium titanate based paraelectric material incorporated Poly (vinylidene fluoride) for biomechanical energy harvesting and self- powered mechanosensing.” Materials Science in Semiconductor Processing, 2023. 153: p. 107128.
Pradeep, D., et al., “An Assessment of the Piezoelectric Coefficient and the Therapeutic Potential of Ionic Liquid (Il) Dissolved Hard Keratin from Goat Horn Discards.” RASAYAN Journal of Chemistry, 2022. 15(04): p. 2914-2921
2024-25:
Sarkar, J., et al., “Roles of aqueous nonsolvents influencing the dynamic stability of poly-(n-butyl methacrylate) thin films at biologically relevant temperatures.” Soft Matter, 2023. 19(42): p. 8193-8202.
Madhusudanan, M. and M. Chowdhury, “An entropy generation approach to the molecular recoiling stress relaxation in thin nonequilibrated polymer films.” The Journal of Chemical Physics, 2024. 160(1): p. 014904.
Madhusudanan, M., et al., “Tuning the Plasticization to Decouple the Effect of Molecular Recoiling Stress from Modulus and Viscosity in Dewetting Thin Polystyrene Films.” Macromolecules, 2023. 56(4): p. 1402-1409.
Jahan, K., et al., “Identification of ethyl-6-bromo-2((phenylthio) methyl) imidazo[1,2- a]pyridine-3-carboxylate as a narrow spectrum inhibitor of Streptococcus pneumoniae and its FtsZ.” (1768-3254 (Electronic)).
Venkatramani, A., A. Ashtam, and D. Panda, “EB1 Increases the Dynamics of Tau Droplets and Inhibits Tau Aggregation: Implications in Tauopathies.” ACS Chemical Neuroscience, 2024. 15(6): p. 1219-1233.
Patra, U., F. Mujeeb, and S. Dhar, “Vapor–Liquid–Solid-Mediated Layer-by-Layer Growth of Stepped-Wedge Shaped WS2 Microribbons Using the Chemical Vapor Deposition Technique.” Crystal Growth & Design, 2024. 24(4): p. 1626-1631.
Piplani, N., et al., “Bulky glycocalyx shields cancer cells from invasion-associated stresses.” Translational Oncology, 2024. 39: p. 101822.
